

- #R studio update column based on another column full
- #R studio update column based on another column code
- #R studio update column based on another column series
As there are 2 entries for o_water these lists have two elements. Here is another approach, in which I extract all the start_ and stop_times of each behavior of o_water into a list. Since the behavioral data is binary, those columns could certainly contain logical data rather than character data, but because they started as character data, keeping them that way seemed simplest. times 7, 8, 12 & 13, in this example) which, once filled in, would make summarizing things like total time spent on each behavior much more straightforward. In this alternate format you could go even further and include time points that weren't explicitly recorded (e.g. Observation_id subject time o_water swim float o_land walk environment Mutate(environment = if_else(o_water = "yes", "water", "land")) # this column is a little redundant now, but here's the water/land column, at last.
#R studio update column based on another column series
#change all "start"s and the first "stop" in each series to "yes", and all other "stop"s to "no" #Then arrange everything in order of time.įill(o_water, swim, float, o_land, walk) %>% Pivot_wider(names_from = "behavior", values_from = "start_stop") %>% # then pivot to w wider format, so each behavior has its own column. Pivot_longer(cols = c("start_time", "stop_time"), names_to = "start_stop", values_to = "time", names_pattern = "(.*)_time") %>% # first pivot to a longer form, so the time values are all in one column Using the data you provided: library(tidyverse)
#R studio update column based on another column code
There's a lot of rearranging that happens in the first few lines I'd suggest stepping through the code one line at a time just to see how each line moves the data around. I'd suggest rearranging it so that each time point has only one record (per individual otter, perhaps), and individual behaviors each have their own column, with binary data indicating whether or not that behavior is occurring at each time point. I think rearranging your dataset will help a lot here. Output observation_id.x subject.x behavior start_time.x stop_time.x environment Select(c(observation_id.x:stop_time.x, environment))
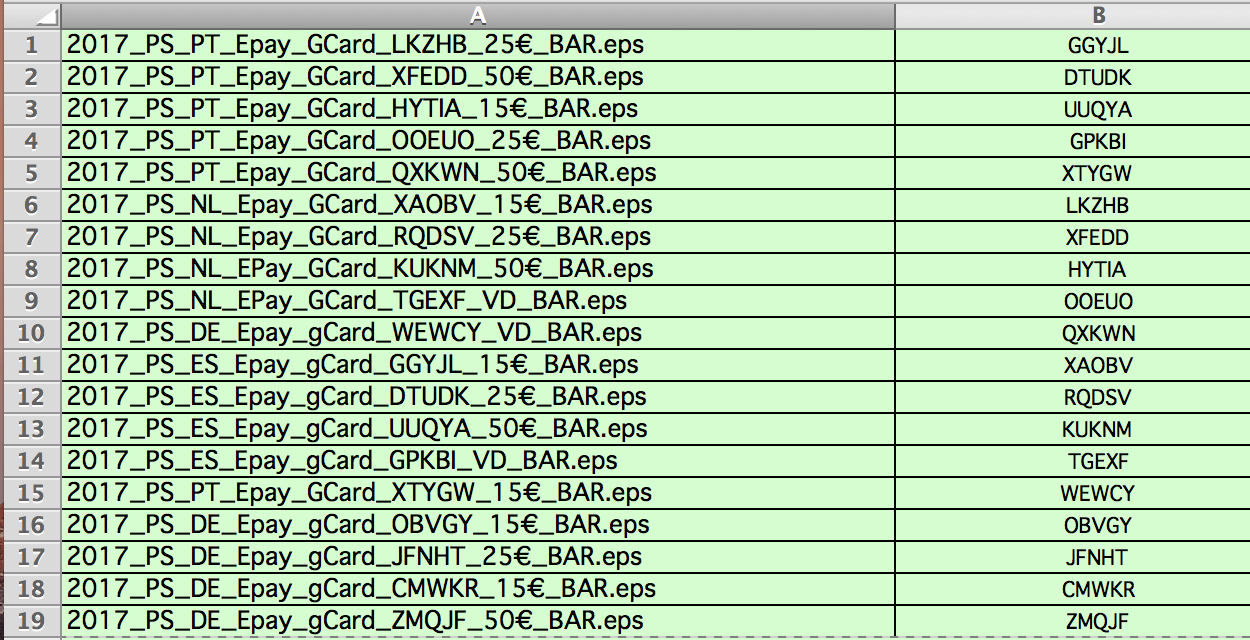
Replace_na(list(environment = "land")) %>% library(dplyr)īy = c("subject", "observation_id", "start_time", "stop_time"), The remaining NA in environment will be non-merged rows, which can be land or other designation. Then, with fuzzy_left_join, merge the o_water rows with the rest of your data, where the start_time and end_time fall between the o_water range. You can separate your o_water behavior rows from otters and designate the environment as water. Here is another approach with dplyr that also uses fuzzyjoin package. I haven't been able to find a problem quite like this elsewhere on stackoverflow. I've tried using when() and case_when() to no avail, but I am very novice level at R so would appreciate any help!Īpologies for any missteps I've done.
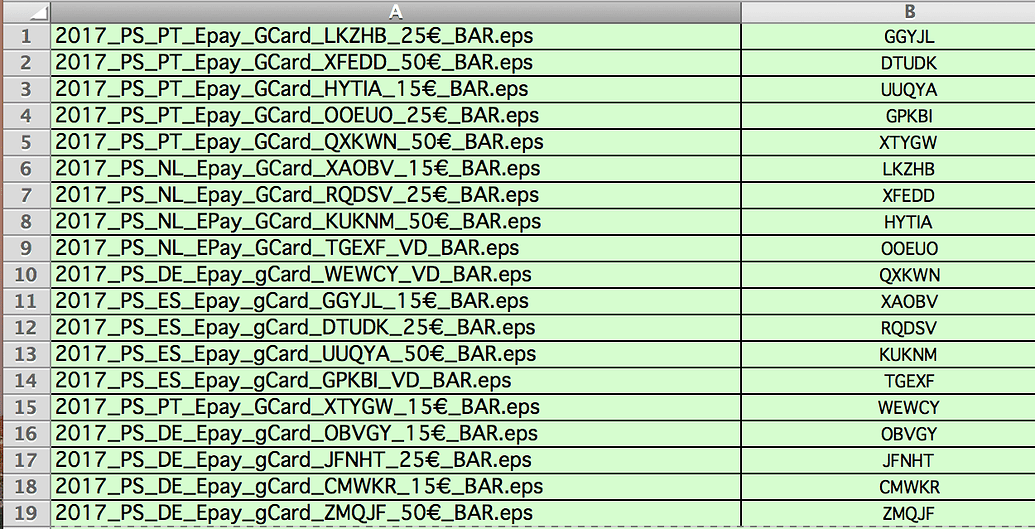
#R studio update column based on another column full
The grouping is so the functions are performed over the applicable rows in the full dataset, there are multiple subjects and observation_id's. The second set of commands is sort of what I want, but I need this to search out and apply it to an entire dataset rather than typing out each parameter. Mutate(environment = ifelse((start_time >= 1 & stop_time = 11 & stop_time Group_by(subject, observation_id, behavior) %>% If this is unclear here is a minimal example: library(dplyr) Specifically, I want the new column to return "water" if the behavior falls between the start/stop times of the behavior "o_water", and "land" if it falls outside these bounds. I am working with a dataset of animal behaviors, and am trying to create a new column ("environment") based on conditions fulfilled in another row.


 0 kommentar(er)
0 kommentar(er)
